This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.
Gibbons Lab Awarded NIH R01 to Explore Precision Nutrition
at the Institute for Systems Biology

Gibbons Lab Awarded NIH R01 to Explore Precision Nutrition
The ecological structure of the human gut microbiome helps determine nutritional and phenotypic responses to diet. However, a clear mapping does not yet exist between diet, gut microbial ecology, gut microbial community metabolism, and human metabolic phenotypes. This proposal, entitled “CyberGut: towards personalized human-microbiome metabolic modeling for precision health and nutrition“, provides five years of funding from the National Institutes of Health (NIH) for the design and testing of an…
Bugs vs. Drugs: How Our Microbiomes Can Explain Our Response to Statins
ISB Assistant Professor Dr. Sean Gibbons talked about the science behind statins in our most recent Research Roundtable virtual presentation. His talk was titled “Bugs vs. Drugs: How Our Unique Gut Microbiomes Shape Our Personalized Responses to Statins.”

ISB Honors Researchers Who Give Back to STEM Education
This year, two deserving scientists were bestowed recognition for giving back to STEM education. Dr. Serdar Turkarslan is the recipient of the JoAnn Chrisman Award for Distinguished Service to STEM Education, and Dr. Christian Diener was awarded the Dr. Christine Schaeffer Award for Exemplary Service to STEM Education.

Interview with Sean in Nature Computational Biology
Dr. Sean Gibbons, assistant professor at the Institute for Systems Biology and a Washington Research Foundation Distinguished Investigator, discusses with Nature Computational Science how he uses computational science to gain insights into the gut microbiome and to address the major challenges of this field, as well as his advice to young LGBTQIA+ scientists.
Gut Microbiome Composition Predictive of Patient Response to Statins
New ISB research shows that different patient responses to statins can be explained by the variation in the human microbiome. The findings were published in the journal Med, and suggest that microbiome monitoring could be used to help optimize personalized statin treatments.

Dr. Annie Levine joins the lab
Dr. Annie Levine joins the lab as a clinical fellow. Annie is a fellow in pediatric gastroenterology at Seattle Children’s Hospital. She graduated from Harvard with a BA in Slavic Languages and Literatures and earned her MD at the University of Pittsburgh in 2017. After completing her residency in pediatrics at Brown University in Providence, RI, she moved to Seattle in 2020 to start her clinical fellowship. Annie has been…
Dr. Jack Gilbert on the State of the Microbiome Field
In the final ISB-Town Hall Seattle Science Series of 2021, ISB Assistant Professor Dr. Sean Gibbons sat down with UCSD Professor Dr. Jack Gilbert, and the two microbiome experts discussed past research, exciting science happening today, promising products and therapies on the horizon, and much more.

2021 ISB Virtual Microbiome Series Announced
Our multi-day microbiome-themed virtual course and symposium is back by popular demand! ISB is hosting a two-day course on October 13 & 14, followed by a symposium on October 15. Both events are virtual and free. The intended audience for these events are graduate students, postdocs, principal investigators, industry scientists, educators, clinicians, or any other variety of microbiome-curious person from across the globe.

Dr. Sean Gibbons Spotlighted in Alumni Profile
MIT’s Center for Microbiome Informatics and Therapeutics recently featured Dr. Sean Gibbons in an alumni profile. Gibbons discussed his academic arc that started in Montana and led to his master’s studies (sponsored by a Fulbright Graduate Fellowship) at Uppsala University in Sweden, completing his PhD in biophysical sciences at the University of Chicago, and conducting his postdoc work in the lab of Eric Alm. He also talked about some of…
Personalized Nutrition and Your Gut Microbiome
In ISB’s first-ever Research Roundtable event, Assistant Professor Dr. Sean Gibbons delivered a presentation titled “Gut-Check: Personalized Nutrition and Your Microbiome.” His talk covered a lot of ground, including recently published research showing how the health of our microbiomes can predict longevity, and how we can build and maintain a healthy gut microbiome.

ISB Research on the Aging Microbiome Featured in The New York Times
ISB’s research into the aging microbiome was featured in a story published by Anahad O’Connor for The New York Times titled “A Changing Gut Microbiome May Predict How Well You Age.” The research featured was published in Nature Metabolism by Drs. Tomasz Wilmanksi, Noa Rappaport, Sean Gibbons and Nathan Price.
Variations in the Microbiome Associated with Health, Disease
ISB researchers examined the associations between the gut microbiomes of about 3,400 people and roughly 150 host characteristics. The team looked at diet, medication use, clinical blood markers, and other lifestyle and clinical factors, and found evidence that variations of the gut microbiome are associated with health and disease.
Answering Nature’s Call: How Scientists Are Mining Sewage To Track Population Health
Everybody pees and poops. What if there was a way to use the byproducts of our everyday bodily functions to understand the general health of a population? That is exactly what MIT’s Dr. Eric Alm is pursuing. In an ISB-Town Hall Seattle live stream, Alm discussed the promise of this novel form of public health tracking.

Nick Bohmann Joins the Lab
Nick Bohmann joins the lab as a Ph.D. student from the Molecular Engineering and Sciences program at UW. Nick graduated from Virginia Tech in 2019 with B.S. in Biological Systems Engineering. His research interests include genome-scale metabolic modeling and ‘omics based computational biology, specifically related to the human gut microbiome. Nick’s work in the lab will focus on using computational tools to enhance the predictive capability of models of the…

James Johnson Joins the Lab
James Johnson joins the lab as a bioengineering PhD student at the University of Washington (UW). James earned his BS in Chemical Engineering from Texas A&M University. After graduating, he took a job as an R&D Associate Engineer at PepsiCo. After three years of applied science and engineering at PepsiCo, James decided to apply to the Bioengineering PhD program at UW in Seattle so that he could return to working…
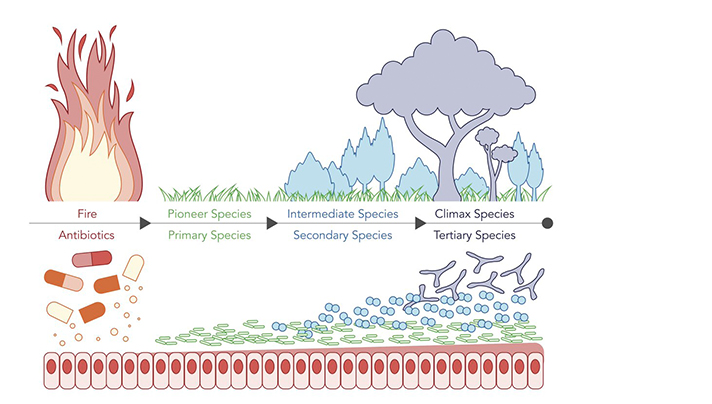
Keystone Taxa Indispensable for Microbiome Recovery
How can we harness successional ecology to quickly repair antibiotic-damaged gut microbiota? ISB Assistant Professor Dr. Sean Gibbons wrote this commentary for the journal Nature Microbiology detailing recent research that answers that question. Click the link to read the story (link will open as a new window). Illustration by Allison Kudla, PhD / ISB.