
Scientists Decode Diet From Stool DNA – No Questions Asked
ISB’s Gibbons Lab developed a breakthrough method that analyzes food-derived DNA in fecal metagenomes, allowing for data-driven diet tracking without the need for burdensome questionnaires.

ISB’s Gibbons Lab developed a breakthrough method that analyzes food-derived DNA in fecal metagenomes, allowing for data-driven diet tracking without the need for burdensome questionnaires.

Reflecting on the past year, ISB has a lot to celebrate: groundbreaking research published in leading scientific journals, well-earned promotions, widespread media coverage, and more. Enjoy our year-in-review roundup highlighting some of the important, interesting, and impactful highlights of 2024.

Alyssa Easton recently joined the lab as a PhD student from the Molecular Engineering Program at the University of Washington. Alyssa is originally from Indiana, where she completed her BS in biological engineering at Purdue University.
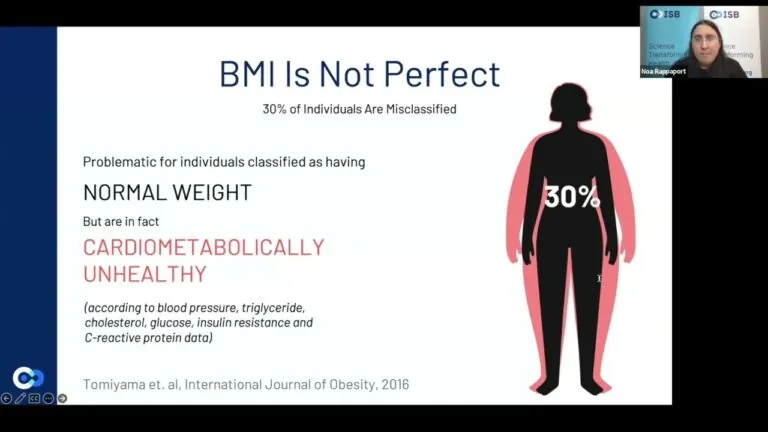
ISB researchers have constructed a biological BMI that provides a more accurate representation of metabolic health and is more varied, informative and actionable than the long-used classical BMI. ISB Senior Research Scientist Dr. Noa Rappaport discussed biological BMI in a Research Roundtable presentation.

ISB researchers have constructed biological body mass index (BMI) measures that offer a more accurate representation of metabolic health and are more varied, informative and actionable than the traditional, long-used BMI equation. The work was published in the journal Nature Medicine.

Gibbons – an expert in microbial ecology and evolution, computational systems biology, the human gut microbiome and its impacts on health, and head of ISB’s Gibbons Lab – has been promoted to Associate Professor.

ISB researchers have shown which blood metabolites are associated with the gut microbiome, genetics, or the interplay between both. Their findings, published in the journal Nature Metabolism, have promising implications for guiding targeted therapies designed to alter the composition of the blood metabolome to improve human health.
The 2022 ISB Virtual Microbiome Series consisted of a two-day virtual course and one-day symposium on global perspectives in microbiome research.